Working with the client we amassed a set of terms that describe neurodegenerative disease in the literature. These terms were used to mine public data to produce gene lists, these lists were expanded into networks by including interaction data. The total set presents a massive network of annotated genes against which omics data for specific diseases can be analysed.
Disease terminology
After a first pass through data sources such as MESH, DO and SNOMED-CT we refined the term list with disease domain experts. These terms were then used to retrieve genes with evidence against those terms from PubMed, ClinVar, GWAS and OMIM (client license).
Data expansion
The large list of candidate genes was expanded by retrieving interacting proteins/genes from Metacore (client license). Annotation was applied to each protein (Uniprot, GO, Interpro).
Data interpretation
Network analysis, statistical analysis of omics data, enrichment analysis and visualisations were performed using R, Cytocape and Pathvisio.
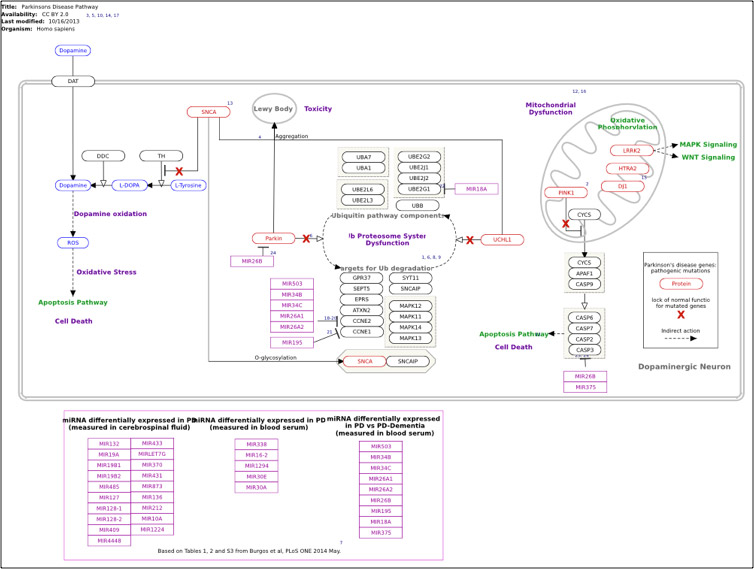
The data package was delivered to the client and is refreshed on a regular basis to facilitate their omics data mining.
