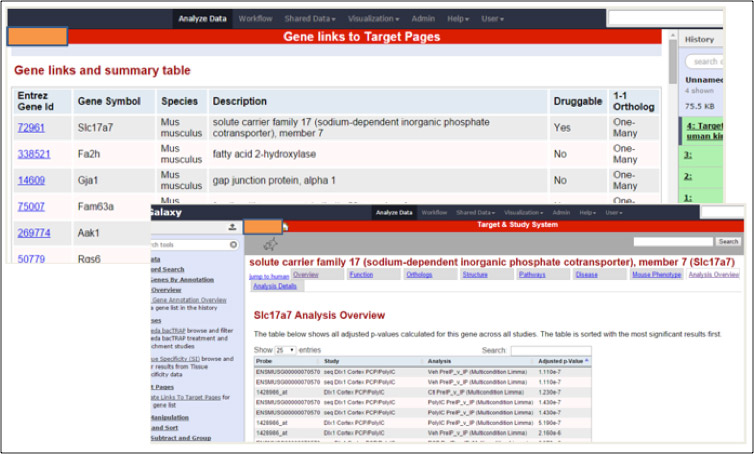
The client generates high-throughput omics data in mice and wants to make that data available to scientists in an easy to access form. However the scale of data production had outstripped the capacity of the existing delivery framework. This project built connected databases for meta-data about omics studies, analysed omics data, and target annotation, so that scientists can ask queries such as “for all GPCR proteins, in which mice experiments are the genes significantly changed in diseases models, and for those is there any disease evidence in humans”.
Annotation Data
Annotation data for human and mouse proteins was sourced from
the GB integrated data system according to client requirements
covering: Function, Pathways, Orthologues, Disease, Structure,
Tissue Specificity and Drug information. That background
annotation was supplemented with client knowledge.
Analysis Data
Here the goal was to capture the output of statistical analysis
of omics data (for instance P-values from a t-test of two
conditions). This was a very complex data problem as omics data
is produced and analysed in studies with different conceptual
configurations. The most similar public domain facilities are
Expression Atlas and GEO Profiles. The system therefore captured
study descriptions using the ISA framework, study configuration,
meta-data about the analyses and stored numeric outputs of the
analyses.
Infrastructure & Deployment
To meet client requirements the database technology is Oracle,
Target summary pages are Java and we deliver bulk querying
functionality via Galaxy.
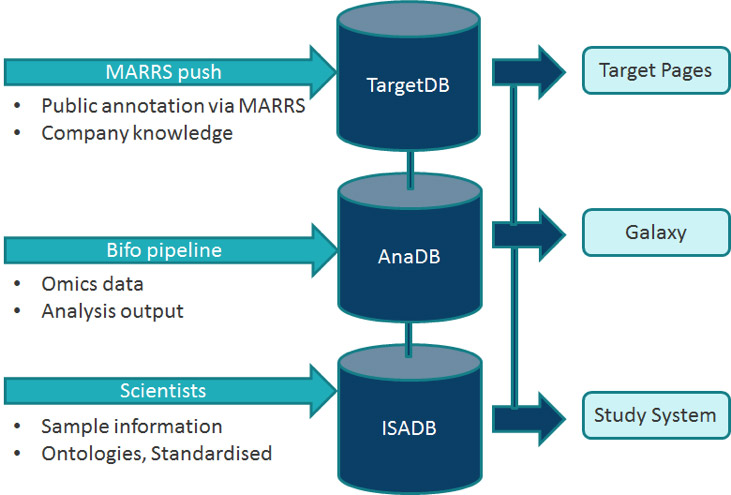
The client now operates the pipeline internally and GB provides troubleshooting and development support. The data focus is primarily transcriptomics (chip and RNAseq) but test data for metabolomics and proteomics has also been included. The project gave all concerned significant experience in the areas of: ISA and associated tools, how best to capture diverse analyses, Galaxy tool development and Transmart; an early candidate for solving this problem that was not pursued.